KEGG pathways extracted using the ROntoTools
R package (update: November, 2021).
References
Kanehisa M, Goto S (1999). KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acid Research 28(1): 27-30. <https://doi.org/10.1093/nar/27.1.29>
Calin Voichita, Sahar Ansari and Sorin Draghici (2021). ROntoTools: R Onto-Tools suite. R package version 2.20.0.
Examples
# \donttest{
library(SEMgraph)
# KEGG pathways
length(kegg.pathways)
#> [1] 225
head(names(kegg.pathways))
#> [1] "EGFR tyrosine kinase inhibitor resistance"
#> [2] "Endocrine resistance"
#> [3] "Antifolate resistance"
#> [4] "Platinum drug resistance"
#> [5] "mRNA surveillance pathway"
#> [6] "RNA degradation"
tail(names(kegg.pathways))
#> [1] "Arrhythmogenic right ventricular cardiomyopathy"
#> [2] "Dilated cardiomyopathy"
#> [3] "Diabetic cardiomyopathy"
#> [4] "Viral myocarditis"
#> [5] "Lipid and atherosclerosis"
#> [6] "Fluid shear stress and atherosclerosis"
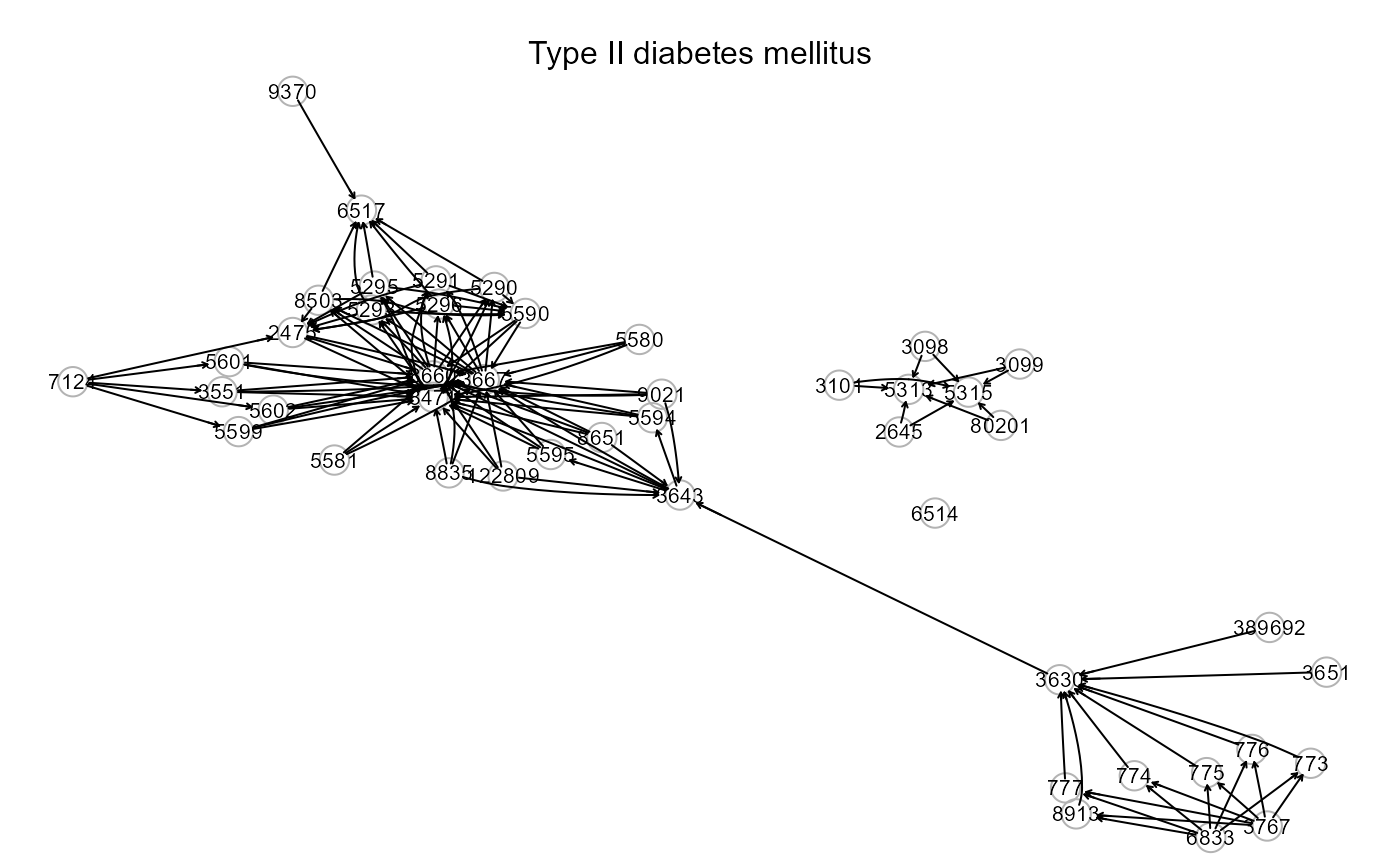
# Load Type II diabetes graph
i<-which(names(kegg.pathways) == "Type II diabetes mellitus");i
#> [1] 127
ig<- kegg.pathways[[i]]
properties(ig)
#> Frequency distribution of graph components
#>
#> n.nodes n.graphs
#> 1 7 1
#> 2 38 1
#>
#> Percent of vertices in the giant component: 82.6 %
#>
#> is.simple is.dag is.directed is.weighted
#> TRUE FALSE TRUE TRUE
#>
#> which.mutual.FALSE
#> 114
#> [[1]]
#> IGRAPH c33de4c DNW- 38 114 --
#> + attr: name (v/c), weight (e/n)
#> + edges from c33de4c (vertex names):
#> [1] 5290 ->6517 5290 ->5590 5290 ->2475 5291 ->6517 5291 ->5590
#> [6] 5291 ->2475 5293 ->6517 5293 ->5590 5293 ->2475 5295 ->6517
#> [11] 5295 ->5590 5295 ->2475 5296 ->6517 5296 ->5590 5296 ->2475
#> [16] 8503 ->6517 8503 ->5590 8503 ->2475 3667 ->5290 3667 ->5291
#> [21] 3667 ->5293 3667 ->5295 3667 ->5296 3667 ->8503 3643 ->3667
#> [26] 3643 ->5594 3643 ->5595 3643 ->8471 3643 ->8660 3630 ->3643
#> [31] 122809->3667 122809->3643 122809->8471 122809->8660 8651 ->3667
#> [36] 8651 ->3643 8651 ->8471 8651 ->8660 8835 ->3667 8835 ->3643
#> + ... omitted several edges
#>
#> [[2]]
#> IGRAPH c33de37 DNW- 7 10 --
#> + attr: name (v/c), weight (e/n)
#> + edges from c33de37 (vertex names):
#> [1] 2645 ->5313 2645 ->5315 3098 ->5313 3098 ->5315 3099 ->5313 3099 ->5315
#> [7] 3101 ->5313 3101 ->5315 80201->5313 80201->5315
#>
gplot(ig, l="fdp", psize=50, main=names(kegg.pathways[i]))
 # }
# }