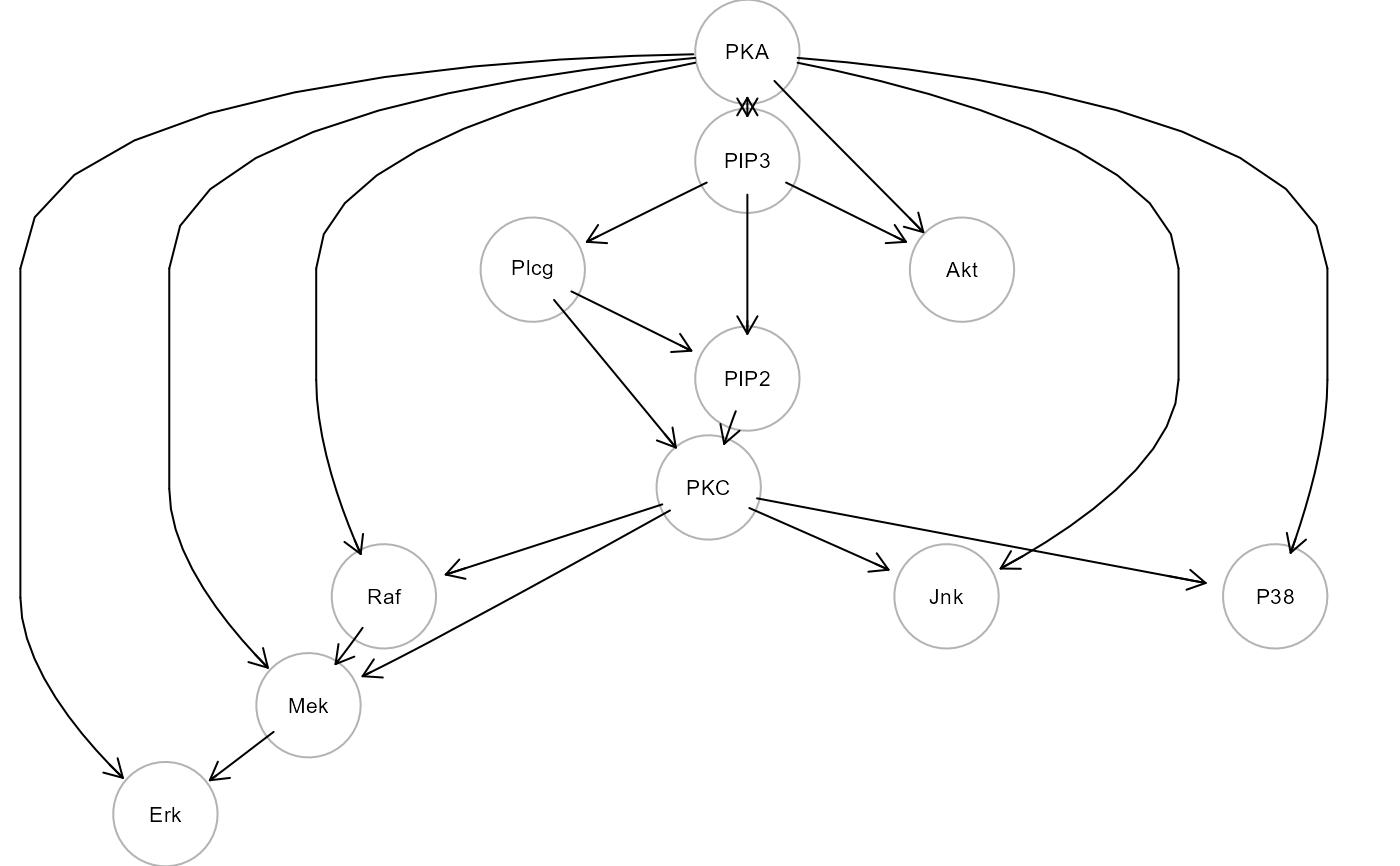
Flow cytometry data and causal model from Sachs et al. (2005).
Format
"sachs" is a list of 5 objects:
"rawdata", a list of 14 data.frames containing raw flow cytometry data (Sachs et al., 2005);
"graph", consensus signaling network;
"model", consensus model (lavaan syntax);
"pkc", data.frame of 1766 samples and 11 variables, containing cd3cd28 (baseline) and pma (PKC activation) data;
"group", a binary group vector, where 0 is for cd3cd28 samples (n = 853) and 1 is for pma samples (n = 913).
"details", a data.frame containing dataset information.
References
Sachs K, Perez O, Pe'er D, Lauffenburger DA, Nolan GP (2019). Causal Protein-Signaling Networks Derived from Multiparameter Single-Cell Data. Science, 308(5721): 523-529.
Examples
# Dataset content
names(sachs$rawdata)
#> [1] "cd3cd28" "cd3cd28_icam2" "cd3cd28_aktinhib"
#> [4] "cd3cd28_g0076" "cd3cd28_psitect" "cd3cd28_u0126"
#> [7] "cd3cd28_ly" "pma" "b2camp"
#> [10] "cd3cd28icam2_aktinhib" "cd3cd28icam2_g0076" "cd3cd28icam2_psit"
#> [13] "cd3cd28icam2_u0126" "cd3cd28icam2_ly"
dim(sachs$pkc)
#> [1] 1766 11
table(sachs$group)
#>
#> 0 1
#> 853 913
cat(sachs$model)
#>
#> # path model
#> P38 ~ PKA + PKC
#> Jnk ~ PKA + PKC
#> Akt ~ PKA + PIP3
#> Erk ~ PKA + Mek
#> Mek ~ PKA + PKC + Raf
#> Raf ~ PKA + PKC
#> PKC ~ PIP2 + Plcg
#> PIP2 ~ PIP3 + Plcg
#> Plcg ~ PIP3
#>
#> # covariances
#> PIP2 ~~ PIP3
gplot(sachs$graph)